DNA Methylation Kits
DNA Methylation is one of the most widely studied epigenetic mechanisms of gene expression regulation. Abnormal changes in DNA methylation have been identified in cancer, inflammatory, autoimmune, psychiatric, cardiovascular, and age-related diseases. Aberrant methylation of increasing numbers of genes is being shown to contribute to diseases such as cancer, diabetes, allergy, Alzheimer's, Parkinson's and more. DNA methylation-based biomarkers are increasingly used in clinical disease management. With this growing field of research, reliable DNA Methylation Testing Kits have become essential. The EpiMelt DNA Methylation Testing Kits are based on MS-HRM technology and provide high accuracy and convenient tests for epigenetics research.
This technology is being used for detection of DNA methylation biomarkers in fields including forensics, and investigating challenging DNA material sources such as liquid biopsies and FFPE tissues.
These kits are fast and robust as well as user-friendly.
See EpiMelt Methylation Detection Workflow here.
See the kit protocol here.
See the SDS for Primer Mix here.
See the SDS for the Controls here.
Epigenetics is a study of the heritable gene expression changes that do not involve alternations in DNA sequence. The prefix epi (meaning on top of or in addition to) has an overall meaning of something on top of genetics that will change the expression of the genes.
The “epi-mechanisms” that influence gene expression are most commonly classified as:
- Covalent modifications of the DNA bases. DNA methylation is the most studied DNA modification leading to gene expression changes.
- Histone modifications: DNA in the nucleus doesn’t exist by itself, but in a complex with proteins called histones. These proteins can regulate access of the transcription machinery to the DNA and hence influence gene expression. This type of gene expression regulation is achieved by covalent modifications of the histones.
- Regulation of gene expression by non-coding RNA mechanisms such as micro-RNA or siRNA.
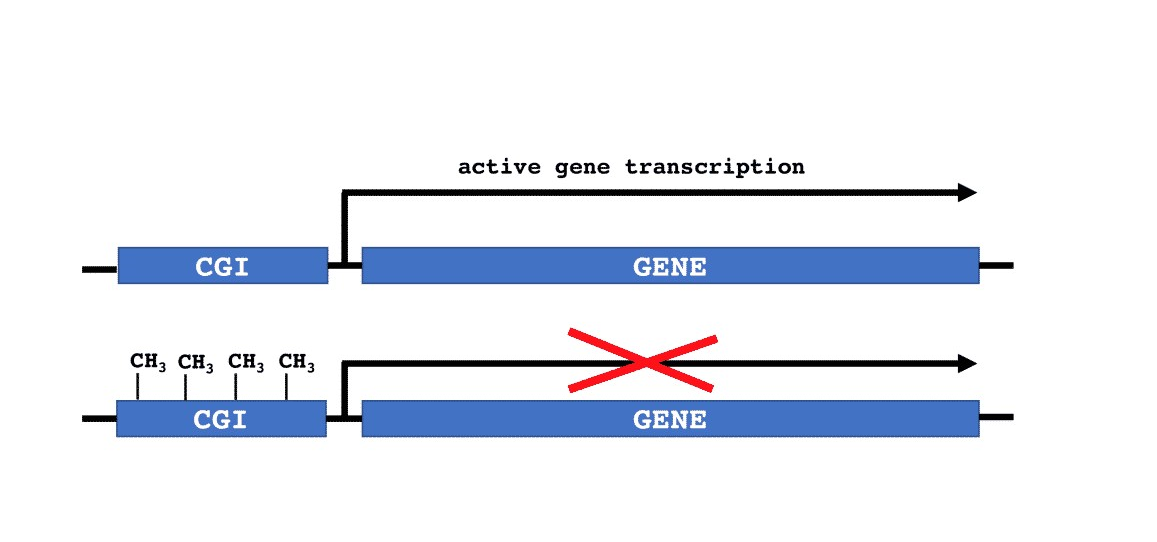
Enzymatic addition of methyl groups (CH3-) to the DNA molecule is referred to as DNA methylation. For mammals, the methyl groups are added to cytosines at CpG dinucleotides (see Figure 1). Specific genomic regions have a high density of CpG dinucleotides and are defined as CpG islands (CGI). When CGIs are located in promoter regions, DNA methylation typically represses gene transcription (Figure 2).
More than 60% of human protein coding genes contain a CGI in the promoter, thus being able to be regulated by DNA methylation. DNA methylation regulates processes such as: imprinting, X-chromosome inactivation, transcriptional repression of transposable elements and aging. Abnormal changes in DNA Methylation have been identified in cancer, inflammatory, autoimmune, psychiatric, cardiovascular, and age-related diseases.
Figure 1
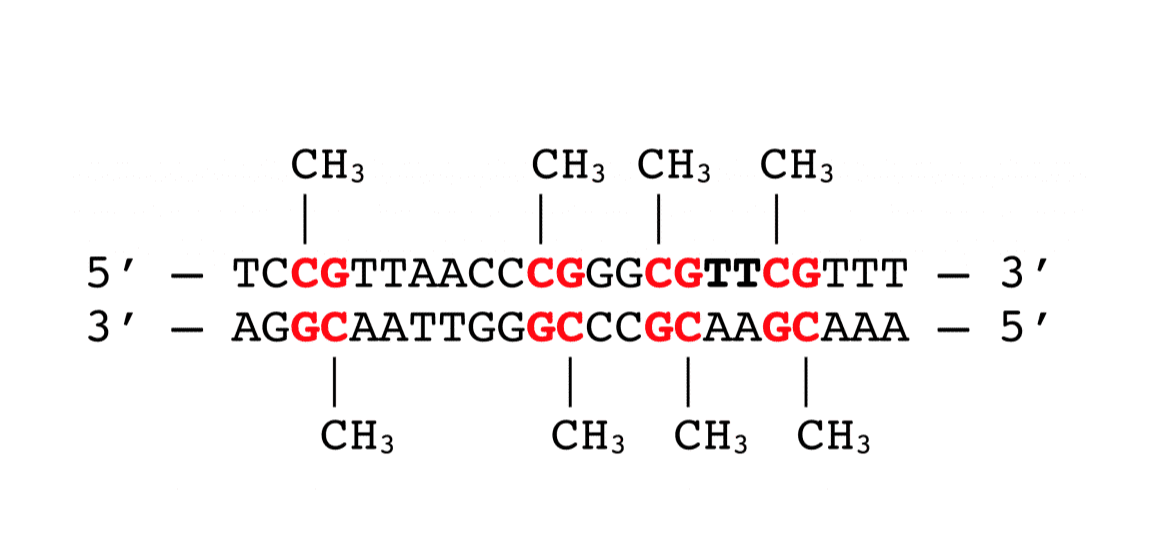
Figure 2
During DNA replication, methyl groups are resynthesized on the newly replicated strand by one of the enzymes called DNA Methyl Transferases (DNMT). Those enzymes are not present in standard PCR reactions. PCR products obtained from the amplification of a specific locus do not contain information about methylation statuses of the cytosines. Therefore, the methylation information is lost. In order to analyze the methylation pattern of cytosines within the locus of interest, the information about cytosines are methylated needs to be preserved before PCR amplification is performed. Sodium bisulfite deaminates non-methylated cytosines to uracil and leaves methylated cytosines untouched. Thus, after bisulfite treatment the cytosine content of the template depends on the number of methylated cytosines in the untreated template (protected from change). PCR products with a different number of cytosines can easily be distinguished in post PCR analyses such as Methylation Sensitive High-Resolution Melting (MS-HRM).
Methylation-Sensitive High-Resolution Melting (MS-HRM) is based on a PCR methylation detection method. The number of cytosines in a PCR product, amplified from a sodium bisulfite modified template, depends on the methylation status of cytosines in the un-modified template. Sodium bisulfite preserves the methylated cytosines and modifies non-methylated cytosines into uracil, which is then substituted with thymine during PCR. Therefore, if the locus is methylated, a C rich PCR product is obtained after PCR amplification, and a T rich PCR product if the locus was non-methylated. When subjected to Methylation-Sensitive High-Resolution melting analyses, these PCR products will display different melting profiles. MS-HRM is dependent on bisulfite modification, which enables cost effective, fast, and highly sensitive assessment of locus specific methylation statuses.
By simply heating double stranded DNA, the dissociation of the double stranded DNA helix occurs, which results in a single strand. This process is known as DNA melting. The temperature at which the DNA strands dissociates into single coils depends on the number of hydrogen bonds holding the complementary strands. There are two hydrogen bonds between adenine (A) and thymine (T) and three bonds between guanine (G) and cytosine (C). Thus, the more G and C present in the sequence, there needs to be a higher temperature in order to melt the DNA fragment.